JMultiPDF.cc File Reference
Example program to test conditional probability density function 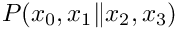
#include <string>#include <iostream>#include <iomanip>#include "TMath.h"#include "TRandom.h"#include "JTools/JHistogram1D_t.hh"#include "JTools/JHistogramMap_t.hh"#include "JTools/JMultiHistogram.hh"#include "JTools/JFunction1D_t.hh"#include "JTools/JMultiFunction.hh"#include "JTools/JMultiPDF.hh"#include "JTools/JToolsToolkit.hh"#include "JTools/JQuadrature.hh"#include "JTools/JQuantile.hh"#include "JIO/JFileStreamIO.hh"#include "JLang/JObjectIO.hh"#include "Jeep/JPrint.hh"#include "Jeep/JTimer.hh"#include "Jeep/JParser.hh"#include "Jeep/JMessage.hh"Go to the source code of this file.
Functions | |
| int | main (int argc, char **argv) |
Detailed Description
Example program to test conditional probability density function 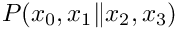
Definition in file JMultiPDF.cc.
Function Documentation
◆ main()
| int main | ( | int | argc, |
| char ** | argv ) |
Definition at line 69 of file JMultiPDF.cc.
70{
72
73 string inputFile;
75 int numberOfEvents;
78
79 try {
80
82
88
89 zap(argc, argv);
90 }
91 catch(const exception &error) {
92 FATAL(error.what() << endl);
93 }
94
95
96 if (numberOfEvents <= 0) {
98 }
99
101
102
106
107
109
112 JHistogramGridMap_t,
113 JHistogramMap_t>::maplist> JMultiHistogram_t;
114
115 JMultiHistogram_t histogram;
116
118
121 for (JGaussHermite::const_iterator x2 = bounds.begin(); x2 != bounds.end(); ++x2) {
122 for (JGaussHermite::const_iterator x3 = bounds.begin(); x3 != bounds.end(); ++x3) {
123 histogram[x0][x1][x0 + x2->getX()][x1 + x3->getX()] = 0.0;
124 }
125 }
126 }
127 }
128
129 // fill
130
132
133 for (int i = 0; i != numberOfEvents; ++i) {
134
135 if (i%1000 == 0) {
137 }
138
139 const double x0 = gRandom->Uniform(xmin, xmax);
140 const double x1 = gRandom->Uniform(xmin, xmax);
141 const double x2 = x0 + gRandom->Gaus(0.0, 1.0);
142 const double x3 = x1 + gRandom->Gaus(0.0, 1.0);
143 const double w = 1.0;
144
145 timer.start();
146
147 histogram.fill(x0, x1, x2, x3, w);
148
149 timer.stop();
150 }
151 STATUS(endl);
152
154 timer.print(cout, true, micro_t);
155 }
156
157 try {
158
160
162
164 }
167 }
168 }
169
170
171 if (inputFile != "") {
172
174 JMAPLIST<JHistogramMap_t>::maplist> JHistogram2D_t;
175
178 JHistogramGridMap_t>::maplist> JMultiHistogram_t;
179
180 JMultiHistogram_t histogram;
181
182 try {
183
185
186 JLANG::load<JIO::JFileStreamReader>(inputFile.c_str(), histogram);
187
189 }
192 }
193
194
197
201
202
203 JMultiFunction_t pdf(histogram);
204
205
206 // test
207
208 JQuantile Q;
209
210 for (int i = 0; i != numberOfEvents; ++i) {
211
212 const double x0 = gRandom->Uniform(xmin, xmax);
213 const double x1 = gRandom->Uniform(xmin, xmax);
214 const double x2 = x0 + gRandom->Gaus(0.0, 1.0);
215 const double x3 = x1 + gRandom->Gaus(0.0, 1.0);
216
217 try {
218
219 const double u = g4 (x0, x1, x2, x3);
220 const double v = pdf(x0, x1, x2, x3);
221
222 Q.put(u - v);
223 }
224 catch(const std::exception& error) {}
225 }
226
227
228 JQuantile V;
229
230 for (JMultiFunction_t::super_const_iterator i = pdf.super_begin(); i != pdf.super_end(); ++i) {
232 }
233
234 cout << "normalisation "
237 cout << "efficiency "
239
242 }
243}
#define make_field(A,...)
macro to convert parameter to JParserTemplateElement object
Definition JParser.hh:2142
General purpose class for multi-dimensional probability density function (PDF).
Definition JMultiPDF.hh:37
void store(const std::string &file_name, const T &object)
Store object to output file.
Definition JObjectIO.hh:68
void load(const std::string &file_name, T &object)
Load object from input file.
Definition JObjectIO.hh:55
This name space includes all other name spaces (except KM3NETDAQ, KM3NET and ANTARES).
Definition JAAnetToolkit.hh:43
JContainer_t::ordinate_type getIntegral(const JContainer_t &input)
Get integral of input data points.
Definition JToolsToolkit.hh:134
Definition JSTDTypes.hh:14
Type definition of a JHistogramMap based on JGridMap implementation.
Definition JHistogramMap_t.hh:36
Type definition of a JHistogramMap based on JMap implementation.
Definition JHistogramMap_t.hh:27
Type definition of a 1st degree polynomial interpolation with result type double.
Definition JFunction1D_t.hh:187
Type definition of a 1st degree polynomial interpolation based on a JGridMap implementation.
Definition JFunctionalMap_t.hh:93
Auxiliary data structure for running average, standard deviation and quantiles.
Definition JQuantile.hh:46
Generated by